21st July 2025 (12 Topics)
Context
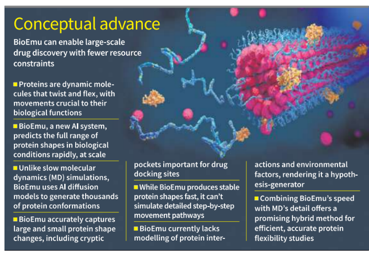
A new deep learning model named BioEmu, developed by Microsoft, Rice University, and Freie Universität, has been unveiled to predict dynamic protein structures more efficiently than traditional methods like molecular dynamics simulations.
BioEmu
Introduction of BioEmu:
- BioEmu is an AI diffusion model trained on:
- Protein structures from AlphaFold predictions
- 200 milliseconds of MD simulations
- 5 million mutant sequences from experimental data
- It uses the reverse-diffusion process to generate equilibrium ensembles — the full range of stable conformations a protein may adopt under physiological conditions.
Key Capabilities:
- Predicts 83% of large shifts and 70–81% of small conformational changes.
- Accurately models local unfolding, cryptic pocket formation, and protein flexibility, even in proteins without a stable 3D structure.
- Enables thousands of conformation predictions in hours on a single GPU.
Applications and Utility:
- Useful in drug discovery, particularly for flexible target proteins such as Ras, implicated in cancer.
- Assists in hypothesis generation for experimental follow-up.
- Can act as a pre-screening tool before running detailed MD simulations.
Limitations:
- Does not model interaction with drug molecules, cellular environments, pH changes, or multi-protein complexes.
- Offers static predictions — unlike MD, it cannot trace step-by-step molecular pathways.
- Prediction reliability scores like those in AlphaFold are not yet integrated.
More Articles